mRNA-peptide fusion
口述:張文章 老師
整理:陳郁鴻
mRNA-peptide fusion
利用linkage每一protein與encode其之DNA sequence(例如mRNA-peptide fusion),做為在in vitro製備nucleic acid-encoded peptide和protein libraries有效率的方法。
在技術方面的考量有別於ribosome display,因為ribosome display過程中可能遭遇ribosome-mRNA-peptide複合體較不穩定的問題,不易於selection時維持其複合體的完整性,因此以peptide與mRNA之covalent linkage的方法在技術上較具優勢。藉由一種抗生素-puromycin,為aminoacyl-tRNA analogue作covalent linkage。
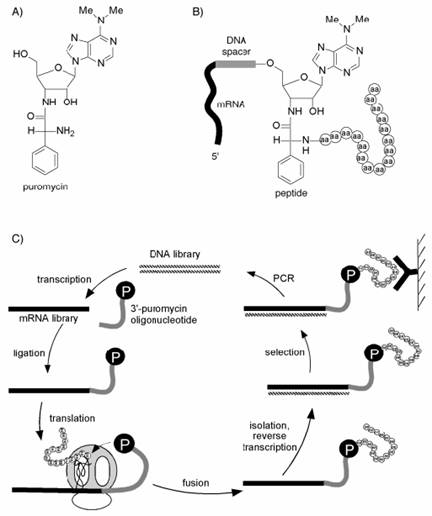
Puromycin分子依附於DNA spacer接在mRNA 3’-end,於in vitro translation時當ribosome停於DNA spacer,末端的puromycin-aminoacyl-tRNA analogue則進入ribosome的A site並與peptide產生linkage,此法亦稱mRNA display。
圖(Figure 7.)顯示典型的mRNA display實驗:
首先將DNA library於in vitro轉錄出mRNA library
↓
合成含puromycin於3’-end之30-bp DNA sequence,並接合至total mRNA
↓
以mRNA-DNA-puromycin hybrid molecular進行in vitro translation,
得以puromycin共價結合之mRNA-peptide複合體
↓
再經isolation、reverse transcription,和利用specific antigen作selection
↓
篩選到的複合體進行PCR放大,可用作定序或轉譯等進行其他實驗
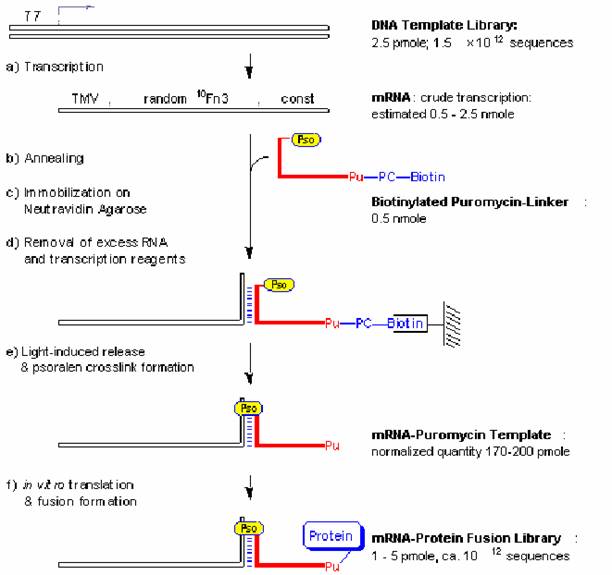
Figure 1:mRNA-protein fusion synthesis
(a) 雙股DNA模板於in vitro轉錄mRNA模板
(b) 與biotinylated puromycin-linker作用
(c) 固定於neutravidin beads
(d) washing並除去unbound mRNA
(e) 將beads照射UV light
(f) elute出photo-ligated mRNA-puromycin template以用來進行in vitro translation構成mRNA-protein fusions library
T7:transcription promoter sequence
TMV:a portion of the tobacco mosaic virus 5’-UTR with good initiation codon context
random 10 Fn3:a sequence coding for a randomized 10th fibronection type III domain
const:a constant sequence which codes for the linker hybridization and crosslinking sites as detailed in Fig. 2
Pu:puromycin
Pso:psoralen
PC-biotin:a photo-cleavable biotin-tag described in Fig. 2.
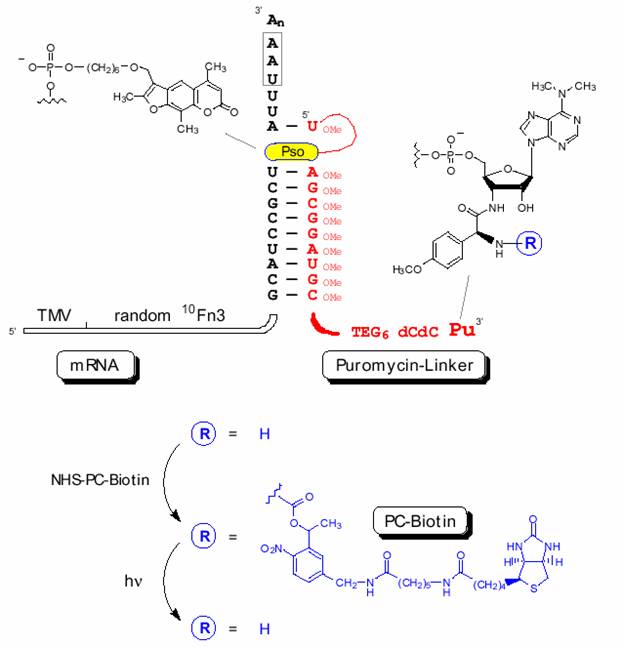
Figure 2:設計之mRNA 3’-end模板和puromycin-linker序列
mRNA-peptide fusion方法的缺點
#所有步驟皆是in vitro情況下進行
#對membrane proteins或必須post-translational modification之proteins則不易操作
參考資料
#Discussion on the paper: H. Lin and V.W. Cornish. Screening and Selection Methods for Large-Scale Analysis of Protein Function Angew. Chem. Int. Ed., 41, 4402- 4425 (2002).
#http://mirror.ensta.fr/molecules/papers/51201259.pdf